Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124

Google Deepmind’s Alphagenomawhich has been announced today is not just another entry into the Ai-For-compiction race. With API access, there is extensive documentation and community support available for non-commercial research-and hosted on Github– Signals that genomics once limited to specialized laboratories and data files are quickly moving towards open science.
That’s quite a big problem.
Imagine your DNA is like a giant instructional guide for how your body works. For a long time, scientists could understand the parts that they told your body to build things like proteins. But most of your DNA – above 90% of it – don’t like it. Nothing builds directly. People called it “unhealthy DNA”.
Now we know that “unsolicited” is actually doing something important: it helps to control when and where real instructions are used – similar to a control panel full of switches and dials. Problem? It’s really hard to read and understand.
There comes alphagenoma.
Alfagenoma is a powerful AI model built by Google Deepmind, which can read these confusing DNA parts better than anything before it. It uses advanced machine learning (such as species for generators or chatbots) to look at huge parts of DNA – except for a million letters long – and find out which parts are important how your genes affect and even how mutations could lead to disease.
It’s something like having a super intelligent microscope AI that not only reads the manual, but also finds out how the system turns on and turns off-and what happens when something goes wrong.
What is fine is that Deepmind shares this tool through the API (a way to talk to computers), so scientists and medical scientists from around the world can use it for free in their research. This means that this could help accelerate discoveries in matters such as genetic diseases, personalized medicine and even anti -aging treatment.
In short: alphagenoma helps scientists to read parts of our DNA, which we have not understood before – and that could change everything about how we treat the disease.
Alfagenoma is a deep learning model to analyze how DNA sequences regulate gene expression and other critical functions. Unlike older models that analyzed short DNA fragments, alphagenoma can process sequences up to one million pairs of basic pairs – an unprecedented scale that allows it to capture remote regulatory interactions by previous methods.
The main power of alphagenoma is a multimodal prediction engine. Unlike previous models that could predict one type of genomic activity, this model publishes high-resolution forecasts for gene expression (RNA-seq, cage), haircut, chromatin conditions (including sensitivity of bottom and histone modification) and 3D chromatin contact maps.
This makes it useful not only for determining which genes are turned on or off in the cell, but also for understanding the complex choreography of the genome of folding, editing and availability.
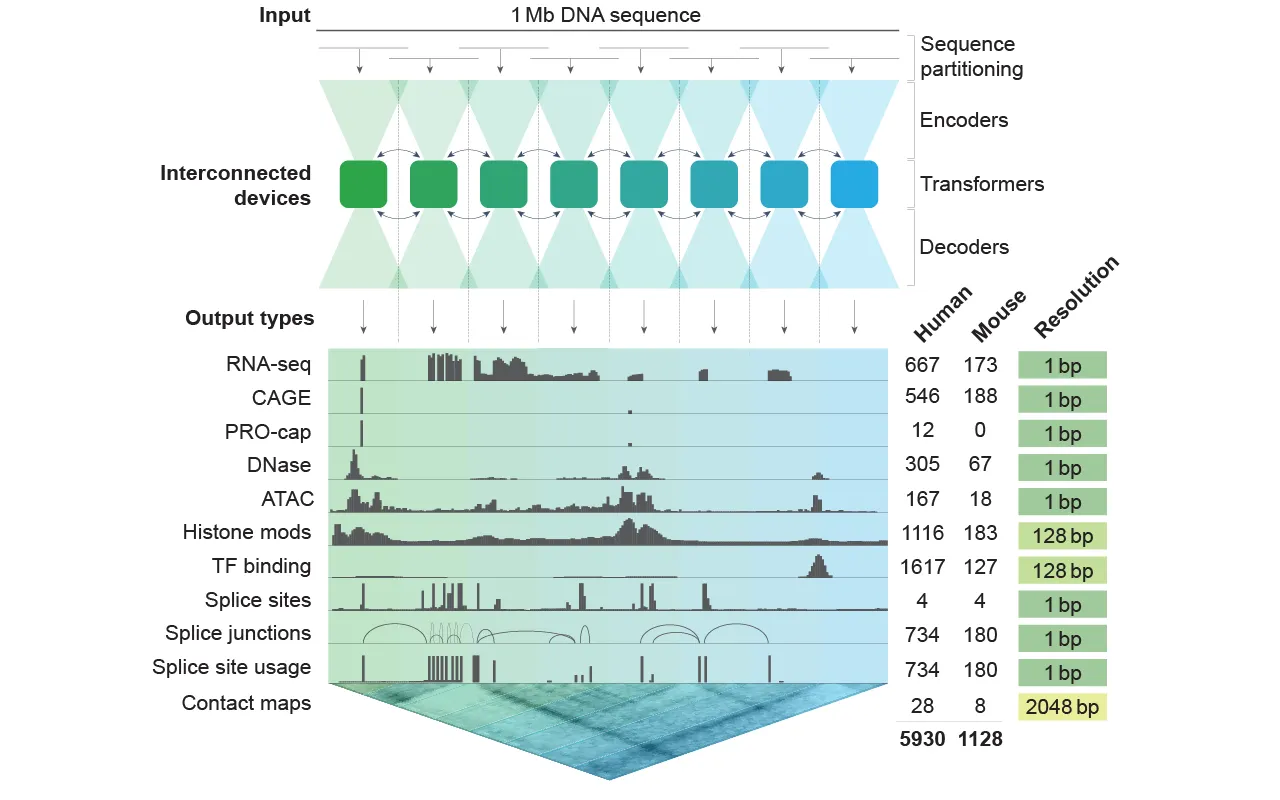
Architecture is remarkable, but still well known if you have used stable diffusion or normal open-source LLM locally: alphagenoma uses a neuron network inspired by the U-NET, with about 450 million trainable parameters.
Yes, this is quite low if you respond against weak and smaller language models that work with billions of parameters. However, since DNA deals with only 4 foundations and only two pairs – primarily the whole human genome is nothing but a combination of 3 billion pairs of AT and CG pairs of letters – it is a very specific model that is designed to do one thing very well.
The model has a sequential encoder, which enters the input from the resolution from one base to the coarser representations, then the transformer model dependence before the decoder reconstructs the output back to the level of one base. This allows predictions for various resolution, allowing both fine and wide regulatory analysis.
The model training relied on a wide range of publicly available data sets, including Concode, GTEX, 4D Nucleom and Fantom5 – lighting, which collectively represent thousands of experimental profiles across human and mice cells.
And this process was also quite fast: using its own Google TPU, the Deepmmation process has completed the process before training and distillation in just four hours using half of the computing budget required by its predecessor, Enformer.
The alphagenoma has exceeded the most modern models in 22 of the 24 prediction tests and 24 out of 26 predictions of variants’ effects, which is a rare clean sweeping in benchmarks, where the standard is increasing improvements. In fact, it does so well that it can compare mutated and unmistakable DNA, predict the impact of genetic variants in seconds – a critical tool for scientists mapping the origin of the disease.
This is important because the non -coding genome contains many regulatory switches that control cell function and the risk of disease. Models such as alphagenoma reveal how much human biology biology is governed by these previously opaque regions.
The influence of AI on biology is difficult to ignore today. Take Ankh, and A model of the protein language He developed teams from the Technical University of Munich, Columbia University and Startup Protinea. Ankh heals protein sequences as a language, generates new proteins and predicts their behavior – similar to the alphagenoma translates the regulatory “Grammar” DNA.
Other neighboring technology, Genslms nvidiaIt shows the ability to predict viral mutations and cluster genetic variants for pandemic research. Meanwhile using AI to promote progress in chemical and gene -based gene Aging intervention It emphasizes the penetration of genomics, machine learning and medicine.
One of the most important contributions of alphagenoma is its availability. Rather than being limited to commercial applications, the model is available through public API for non -commercial research.
Although it is not yet fully open – that is, researchers cannot download and operate or modify locally – API and accompanying resources allow scientists to generate predictions around the world, adapt analyzes for different types or cell types, and provide feedback for forming future editions. Deepmind signaled plans for a wider open-source release by line.
The ability of alphagenoma to analyze non-coding variants-containing, where most mutations associated with diseases-by could unlock a new understanding of genetic disorders and rare diseases. Its high -speed scoring also supports personalized medicine, where treatment is adapted to the unique DNA profile of the individual.
For the time being, the non -coding genome is less black box and the role of AI in the genomic is only set to expand. Alfagenom does not have to be a model that takes us to Huxley’s “Brave New World”, but it’s clear signs of where things are going: more data, better predictions and a deeper understanding of how life works.
Weekly journey AI narrated gene, generative model AI.




